Molecular characterization of abortive infections
Abortive infections of permissive cells are abundant but have not been mechanistically studied due to the lack of appropriate tools. Our lab is testing the hypothesis that activation of specific host transcriptional programs is able to abort infection, even after viral gene expression has begun.
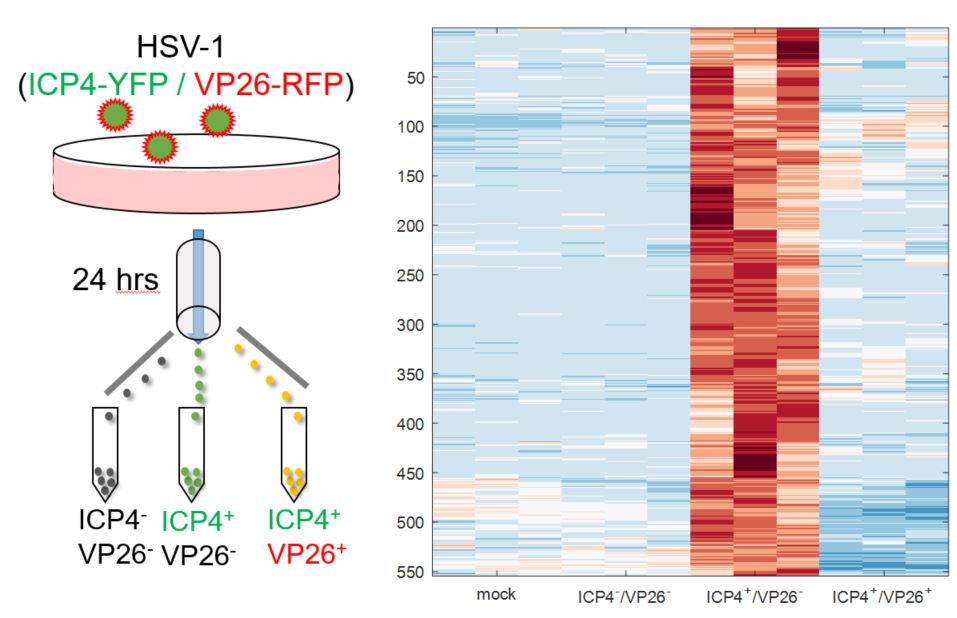
To visualize abortive infections by HSV-1, we created a new viral strain which harbors two fluorescently-tagged proteins; ICP4-YFP and VP26-RFP. ICP4 is an immediate-early gene, expressed as soon as the viral genome enters the nucleus while VP26 is a late gene, expressed only after viral DNA replication.
Using this strain, three subpopulations of infected-cells are seen at 24 hours post-infection: non-infected (ICP4–/VP26–), enriched for abortive infections (ICP4+/VP26–) and enriched for productive infections (ICP4+/VP26+). These three populations, as well as mock-infected cells, were collected by FACS and profiled by RNA-sequencing, identifying 554 host genes that are specifically up-regulated in abortively-infected cells . Many of these are uncharacterized and potentially represent novel antiviral signaling pathways.
What makes a cell predispose to become a super producer?
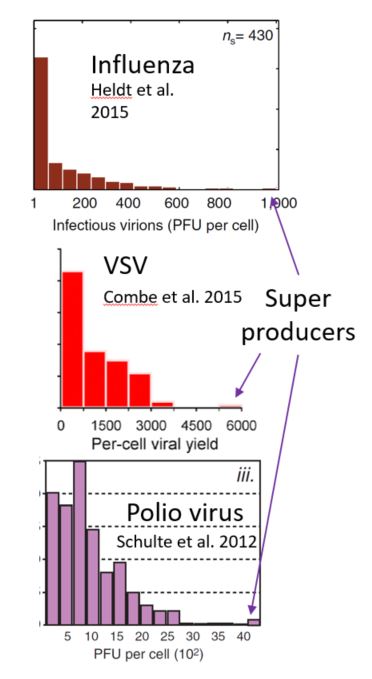
In the few cases it has been quantified, progeny production by individual cells showed a long tail distribution, in which most infected cells produce few progeny, but a small subset of cells produce thousands. Two observations suggest a critical role for host cell factors in shaping the super producer phenotype: (1) the distribution of progeny production by individual cells did not depend on the MOI and (2) the amount of viruses produced from individual cells was not a heritable trait (meaning viruses obtained from individual cells producing low or high amounts of progeny produced similar amounts of progeny upon re-infection).
Currently, the cellular events that shape a cell’s fate as a super producer are completely uncharacterized, due to technical limitations in characterizing and studying these cells. Our lab will test the hypothesis that specific cellular pathways are activated in cells that become super producers by developing computer-vision tools to connect the image of a cell before infection with the number of progeny it produces. This will allow us to identify cells that will become super producers, using the cell’s image before infection. These cells will then be analyzed to reveal the cellular states that support super production.
Discovery and optimization of antiviral compounds
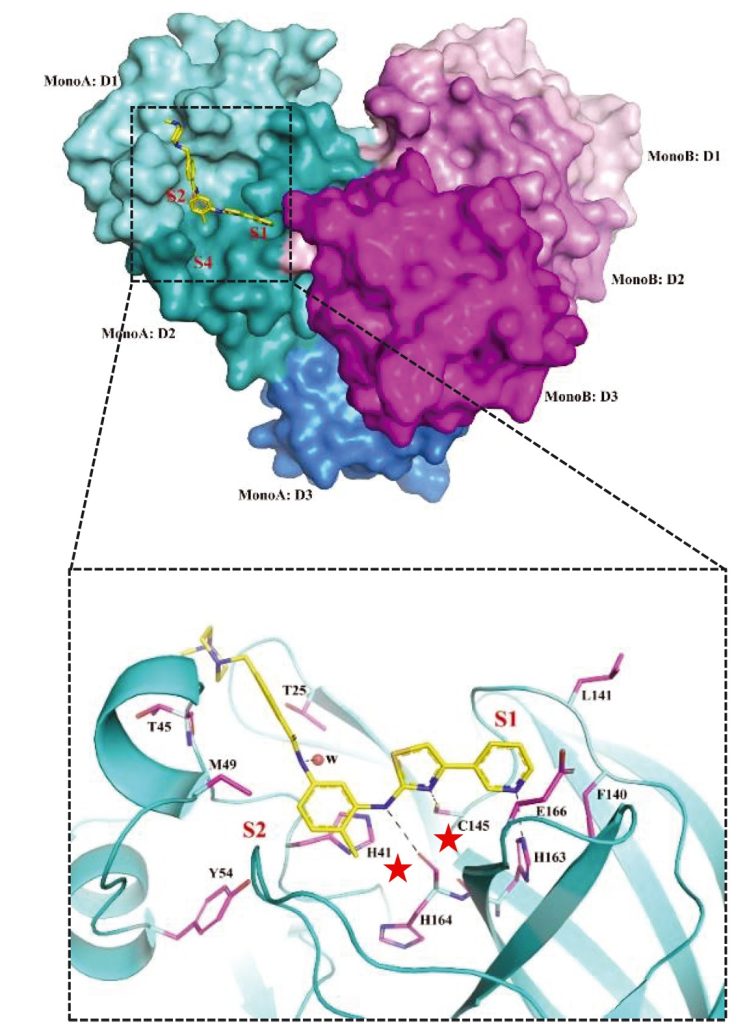
During my post-doctoral studies I screened a library of 1,900 safe-in-human drugs for their ability to inhibit the replication of coronaviruses and identified masitinib, an orally bioavailable c-kit inhibitor in multiple phase 3 clinical trials, as a pan-coronavirus inhibitor (Drayman et al, Science 2021). We found that masitinib inhibits multiple coronaviruses by inhibiting the activity of their main protease (3CLpro) and used x-ray crystallography to show masitinib directly binds and blocks the protease active site. Masitinib is currently being tested in Phase 2 clinical trials to test its effect on COVID-19 patients.
Our lab will collaborate with the company that manufactures masitinib (AB Science, France) to design better antivirals, using masitinib as a backbone.